A Flocking Algorithm using OpenGL and Parallel Computing Libraries
Bees swarm, fish school and birds of a feather flock together. Especially those geese I see flying south every winter! But how do they manage such feats?! Believe it or not it is possible to model the behavior of flocking animals in code!
In 2019 I implemented a flocking algorithm in C++ using OpenGL and tested out it’s performance using parallel computing tools as a project. I was really impressed with the performance I got and had a lot of fun exploring flocking algorithms.
The basic principle of the algorithm is that there are individual agents, called “boids”, that represent the animals in a flock.
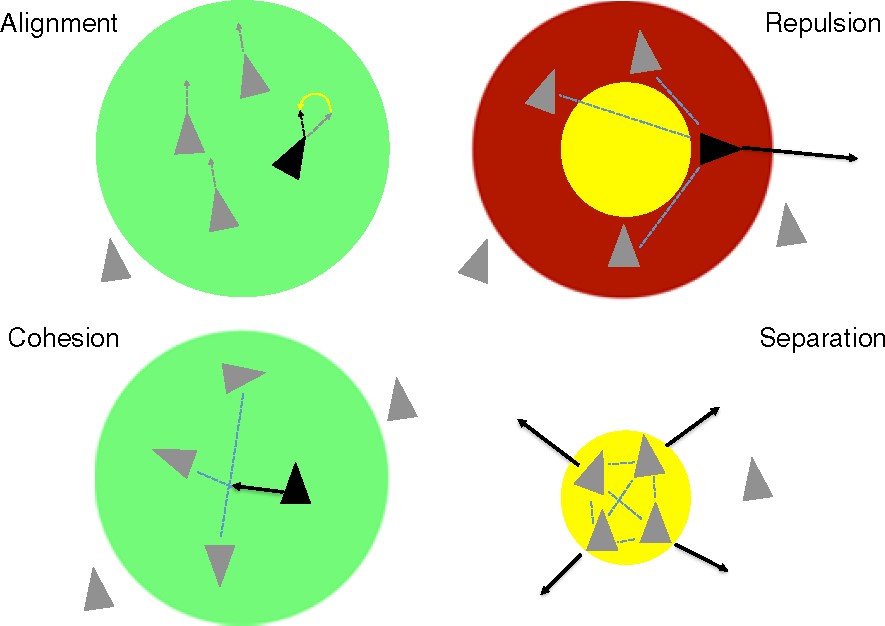
Boids in a flock can follow many different rules. The following are the four rules I utilized in my project, but there certainly could be others in a more complex model:
Avoidance: Boids move away from other boids to avoid collisions.
Copy: Boids fly in approximately the same direction as the rest of the flock by averaging velocities and directions of travel.
Center: Boids try to move toward the center to stay out of the fringes.
View: Boids try to change their position and velocity so that other boids do not blocktheir field of view.
So, in each iteration of the main loop in the code, each of the rules gets updated for each boid and thus the flocking behavior is simulated!
Interestingly enough, these rules might apply to a variety of situations in the real world.
Narechania, et al. even applied the flocking algorithm to an analysis of phylogenomic datasets in 2016 which seems likely a surprisingly unrelated scenario but nonetheless they found thatconceptually the flocking algorithm suited their research challenge nicely (see Fig. 1). In nature, of course, flocking scenarios abound. Everything from birds, to fish, to ants and even many insects flock together in groups, though the rules might differ for different organisms. In fact,just by modifying the relative contributions of the four rules above we can obtain different emergent behaviors for our virtual flock and get it to behave more like Canada Geese, or a swarm of gnats, or a shoal of fish.
The narechania paper link has a nice illustration of how this works:
The final simulation I was able to produce, using C++ and OpenGL, has a flock of boids swimming inside of 3D model of an aquarium:

The only drawback of this simulation is that it runs really slow when you add a lot of boids. Even a hundred boids can take a lot of processing power to simulate, because the rules need to be updated for each boid on each iteration.
Here’s where parallel computing comes to the rescue!!!
Using APIs for parallel processing in C++ I was able to greatly improve the performance of the flocking simulation simply by allowing multiple cores to calculate all the rules!
Both OpenMP and MPI proved to be really useful for this. OpenMP link is a freely available high-level multithreading parallelism API available for Fortran and C/C++. MPI link is a similar API that is designed around message passing. They both have different advantages and can even be used together (e.g. on a computer cluster), to take maximum advantage of available computing resources.
Parallelizing with OpenMP
For the OpenMP version the code for applying the rules was modified essentially as such:
1 | #pragma omp parallel shared(theFlock) private(j) |
This directed OpenMP to use the flock class as a shared variable and the loop index of the innerloop as a private variable for each process. So each time a boid is processed different iteration of the inner loop are assigned to distinct processors. A barrier is needed at the end to prevent raceconditions. The pragma directives are the primary way of integrating OpenMP into the code.
Parallelizing with OpenMP
For the MPI version I modified the same region of code.
Parallelizing with MPI was somewhat more involved because the serial code stored boids as class objects using C++ class definitions but MPI standards only allow for datatypes that are declared in a way that MPI can understand the memory structure of the data, so the boid data had to be reorganized into C style structs and a special MPI datatype needed to be declared.This was done by calculating the memory offsets of data items in the boid data struct and then calling MPI_Type_create_struct as well as MPI_Type_commit with this new data type information.
The pseudocode for the MPI implementation looks roughly like this with message passing directives indicated:
1 | While Not Done |
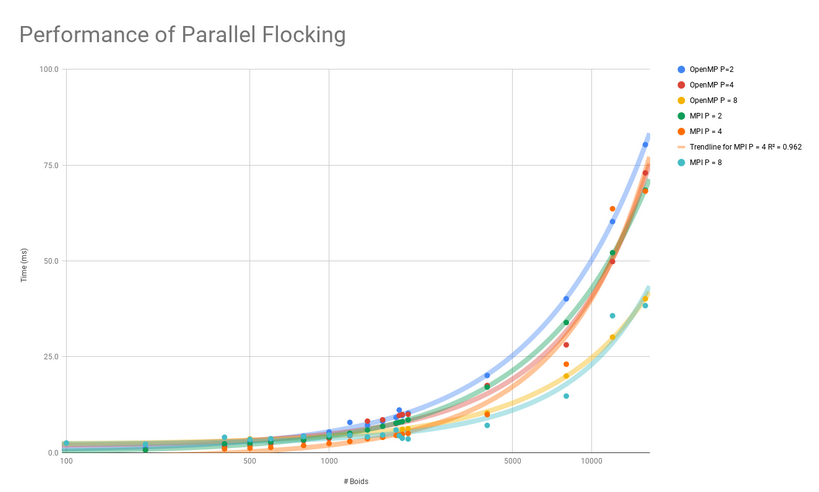
Performance:
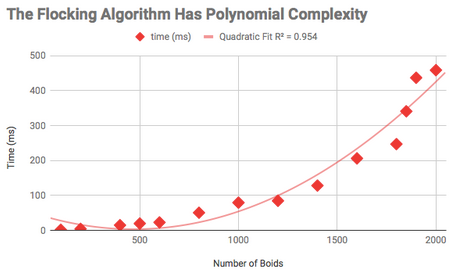
The parallel versions of my code ran a lot faster and allowed me to simulate many more boids than before. With the serial version I could simulate up to 2,000 boids at once but at that point it took about half a second to update all the rules on each iteration. This was really slow. But with the parallel APIs I could simulate more than 15,000 boids at once on the same desktop computer substantially faster! Both MPI and OpenMPI allowed me to simulate about 15,000 boids at less than 100ms per iteration, which was a huge speed-up.
Overall I would say this was a huge success. These parallel computing tools were relatively easy to add to the code and the performance improvements were huge. For tasks such as this where parallelization is possible these APIs are excellent choices.
I’ve made some plots of the performance data for anyone interested below:
Serial Version Performance
Parallel Performance